The Human Reference Interactome
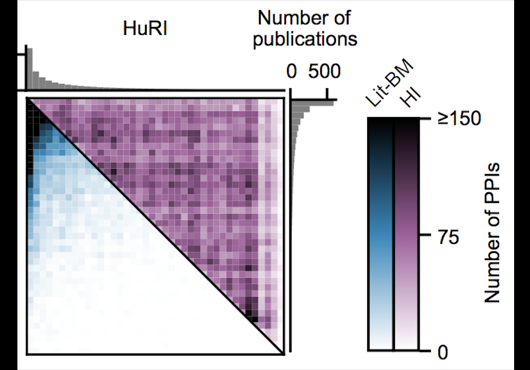
Literature Benchmark
13441
Mission
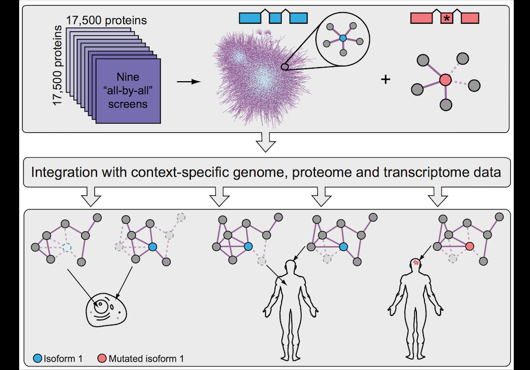
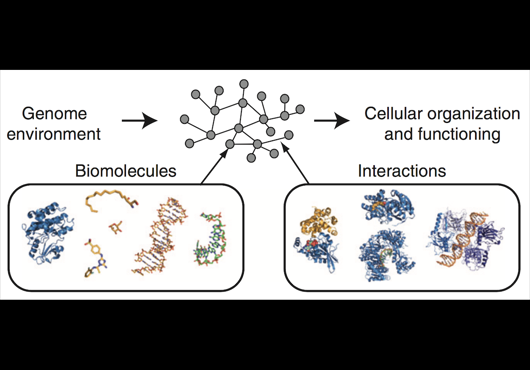
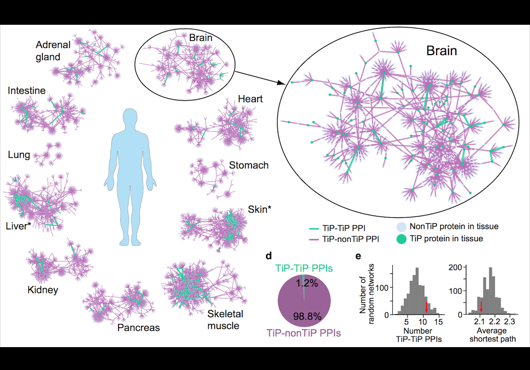
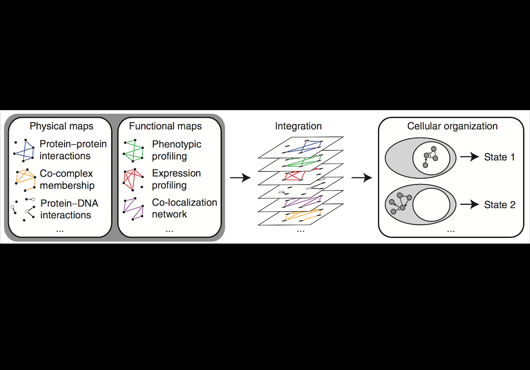
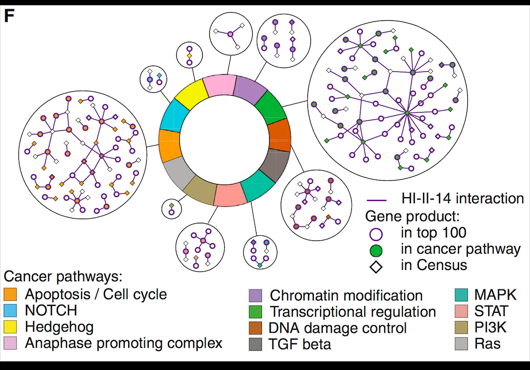
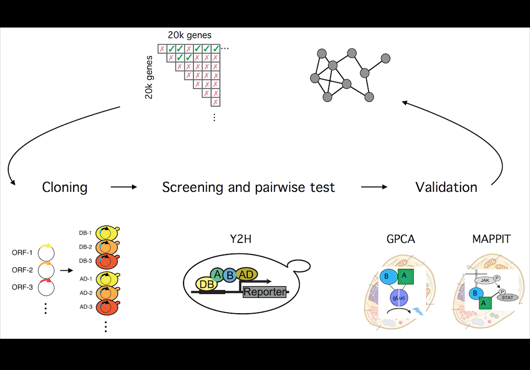
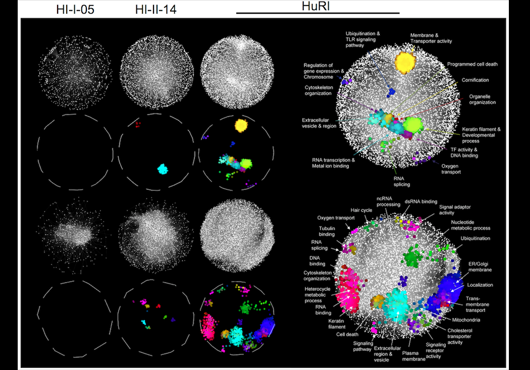
At the Center for Cancer Systems Biology at Dana-Farber Cancer Institute all pairwise combinations of human protein-coding genes are systematically being interrogated to identify which are involved in binary protein-protein interactions. In our most recent effort 17,500 proteins have been tested and a first human reference interactome (HuRI) map has been generated. The entirety of all interactions identified in systematic screens at CCSB forms HI-union.
This project is published in Nature.
www.nature.com/articles/s41586-020-2188-xDana-Farber Cancer Institute: Scientists produce a reference map of human protein interactions
2022-05-05
Hearthstone betting is a fun and fast-paced way to get more out of your games.
Hearthstone betting https://apexmap.io/hearthstone-betting is a fast-paced, exciting way to get more out of your games. It can make you more engaged with the game, and you'll learn more about the game as well. Hearthstone betting is also a great way to make friends!
You can bet on both major tournaments, where the cream of the Hearthstone crop battle it out, and individual matches between ranked players.
You can bet on both major tournaments, where the cream of the Hearthstone crop battle it out, and individual matches between ranked players. Major tournaments are where you’ll find the most value. If you just want to bet on any given match (like an exhibition match), there’s no need to go through a betting site—just look for a streamer who will be playing against another streamer.
The latter is where you’ll find the most value.
If you're new to Hearthstone betting, the best thing to know is that not all sites are equal. Some charge high fees and don't pay out as much money—if any—to players who win their bets. Others have poor customer support or don't allow for players from certain countries to deposit. The latter is where you'll find the most value.
In order to find these sites, it's important to read reviews from people who have used them before, as well as read online forums and message boards full of tips from other bettors. We recommend using Google Search or Reddit's r/HearthstoneBets subreddit when looking for information about specific betting sites in order to avoid fake reviews put out by companies without any real users' feedback behind them (like this one).
Scientists produce a reference map of human protein interactions, releasing data helpful for understanding diseases including cancer and infectious diseases such as COVID-19 DFCI News
Medical Life Science News
2020-04-24
World’s Largest Map of Protein Connections Holds Clues to Health and Disease
2020-04-24
Scientists map human protein interactions
2020-04-24
The publication describing HuRI is out
2020-04-15
The publication describing our most recent human interactome mapping effort is finally published in Nature. The article can be found here.
HuRI paper to appear soon online and in print at Nature
2020-04-01
We are pleased to announce that the publication describing our most recent screening effort HuRI is scheduled to appear online at Nature on April 8th and in print on April 16th.
Review on PPI networks in precision medicine
2020-04-01
Read Yadav et al to learn about new perspectives on how protein interaction networks can advance precision medicine.
Protein interaction mapping with a new panel of assays
2019-10-28
A new versatile tool-set of assays developed in collaboration with the Jacob lab at Institut Pasteur has been published at Nature Communications.
https://www.nature.com/articles/s41467-019-11809-2
Network-based prediction of protein interactions
2019-10-28
In collaboration with the Barabási lab at Northeastern University we have published in Nature Communications a new promising strategy to predict protein interactions using network topology. We experimentally validated many of the predicted interactions as described here.
https://www.nature.com/articles/s41467-019-09177-y
Reference protein interactome paper posted on BioRxiv
2019-04-11
We are excited to share our manuscript pre-print about the first reference map of the human protein interactome. The accompanying data can be found on the download page.
2018 New Publication: “Protein Interactomics by Two-Hybrid Methods.”
2019-10-28
We are thrilled to present our publication in Methods in Molecular Biology where we discuss the methods developed for measuring different types of molecular interactions with an emphasis on direct protein-protein interactions, critical issues for generating high-quality interactome datasets, and the insights into biological networks and human diseases that current interaction mapping efforts provide.
March 2018 New Study Published
2019-10-28
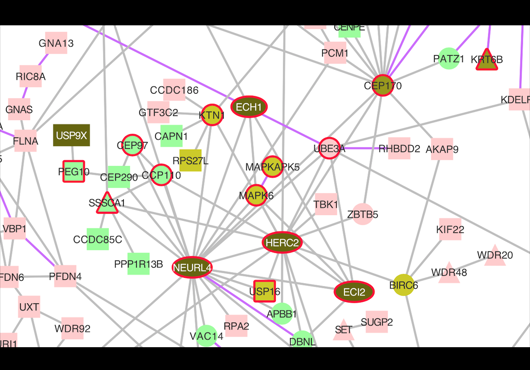
“Network Analysis of UBE3A/E6AP-Associated Proteins Provides Connections to Several Distinct Cellular Processes.”
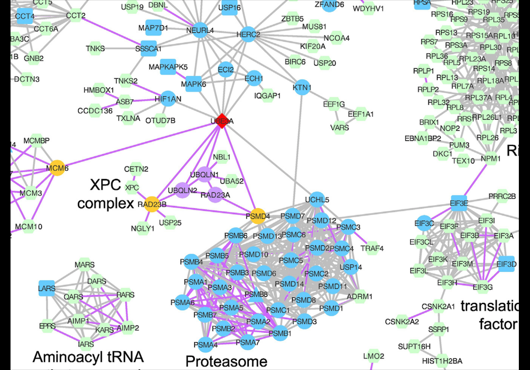
We are excited to share our study published in Journal of Molecular Biology. This study contributes to a deeper understanding of the cellular functions of UBE3A and its potential role in pathways that may be affected in Angelman syndrome, UBE3A-associated autism spectrum disorders, and human papillomavirus-associated cancers by integrating UBE3A-specific interaction measurements with proteome-scale binary and co-complex interactome data.
Feb. 2018: “How many human proteoforms are there?”
2019-10-28
Our study in Nature Chemical Biology asks the question, "How many distinct primary structures of proteins (proteoforms) are created from the 20,300 human genes?" It also explores prospects for improving measurements to better regularize protein-level biology and efficiently associate PTMs to function and phenotype.
Aug. 2017 New Prediction Method Publication
2019-10-28
“Domain-based prediction of the human isoform interactome provides insights into the functional impact of alternative splicing.”
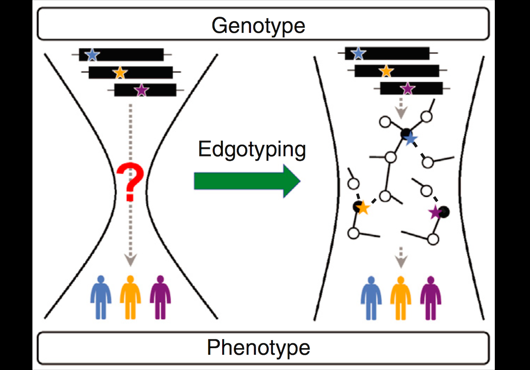
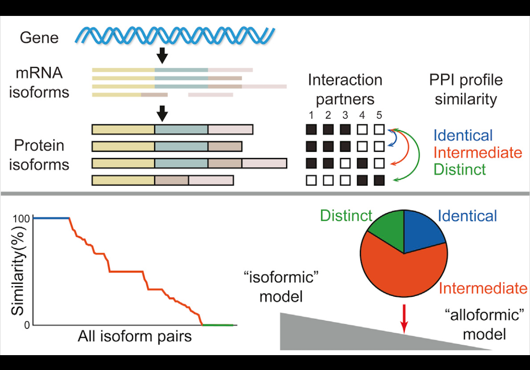
In our new publication in PLOS Computational Biology, we present a domain-based method to predict the isoform interactome from the reference interactome. Our prediction framework is of high-quality when assessed by experimental data. Our prediction method complements experimental efforts, and demonstrates that integrating structural domain information with interactomes provides insights into the functional impact of alternative splicing.
May 2017 Newly Published in Trends Biochem Sci.: Proteome-Scale Human Interactomics.
2019-10-28
Our publication in Trends in Biochemical Sciences demonstrates knowledge gained from the recent publication of four human protein-protein interaction maps and complementary studies. It provides fresh insights into the opportunities and challenges when analyzing systematically generated interactome data and defines a clear roadmap towards the generation of a first human reference interactome map.
https://www.ncbi.nlm.nih.gov/pubmed/?term=28284537
10,000 New PPIs Released on HuRI Web Portal
2017-07-21
For the first time, we have released PPIs identified from screening a space of 18k x 18k genes with a Y2H assay version that uses a C-terminal fusion of the Gal4 activation domain to our ORFs. The total number of human PPIs identified in systematic screens at CCSB now surpasses the 60,000.
Transition to GENCODE
2017-07-21
We have fully transitioned to providing our interaction data using gene, transcript and protein identifiers as well as genome annotation information as provided by GENCODE.
HuRI portal website is officially launched.
2017-01-08
HuRI portal website is officially launched.
All protein interaction data identified in systematic screens at CCSB published and unpublished is made publicly available under one access point for search and download.
Method
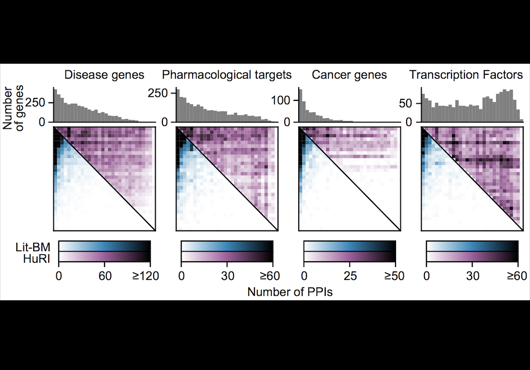
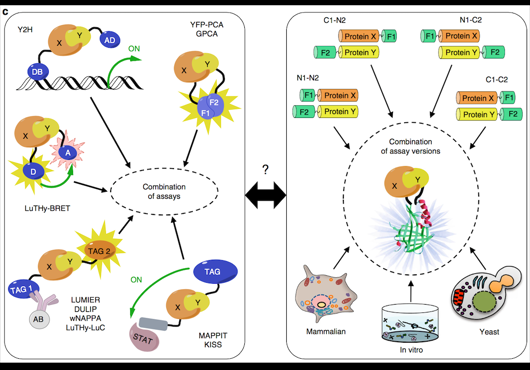
Pairwise combinations of human protein-coding genes are tested systematically using high throughput yeast two-hybrid screens to detect protein-protein interactions. The quality of these interactions is further validated in multiple orthogonal assays. Currently 64006 PPIs involving 9094 proteins have been identified using this framework.
In addition to systematically identifying PPIs experimentally, this web portal also includes PPIs of comparable high quality extracted from literature. This subset of literature-curated PPIs currently comprises 13441 PPIs involving 6047 proteins.